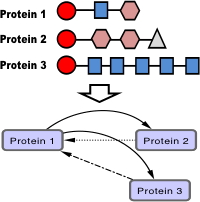
MOCASSIN-prot
Multi-Objective Clustering Algorithm for Sequence Similarity Networks for proteinsKeel, B. H., Deng, B. and Moriyama, E. N. (2018) MOCASSIN-prot: A multiple objective clustering approach for protein similarity networks. Bioinformatics 34: 1270-1277.
AGIdb
Arabidopsis G-signaling Interactome DatabaseKlopffleisch, K., Phan, N., Augustin, K., Bayne, R. S., Booker, K. S., Botella, J. R., Carpita, N. C., Carr, T., Chen, J.-G., Cooke, T. R., Frick-Cheng, A., Friedman, E. J., Fulk, B., Hahn, M. G., Jiang, K., Jorda, L., Kruppe, L., Liu, C., Lorek, J., McCann, M. C., Molina, A., Moriyama, E. N., Mukhtar, M. S., Mudgil, Y., Pattathil, S., Schwarz, J., Seta, S., Tan, M., Temp, U., Trusov, Y., Urano, D., Welter, B., Yang, J., Panstruga, R., Uhrig, J. F. and Jones, A. M. (2011) Arabidopsis G protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Molecular Systems Biology 7: 532.
7TMRmine
A hierarchical mining tool for 7TMRsLu, G., Wang, Z., Jones, A. M., and Moriyama, E. N. (2009) 7TMRmine: A Web server for hierarchical mining of 7TMR proteins. BMC Genomics 10: 275.
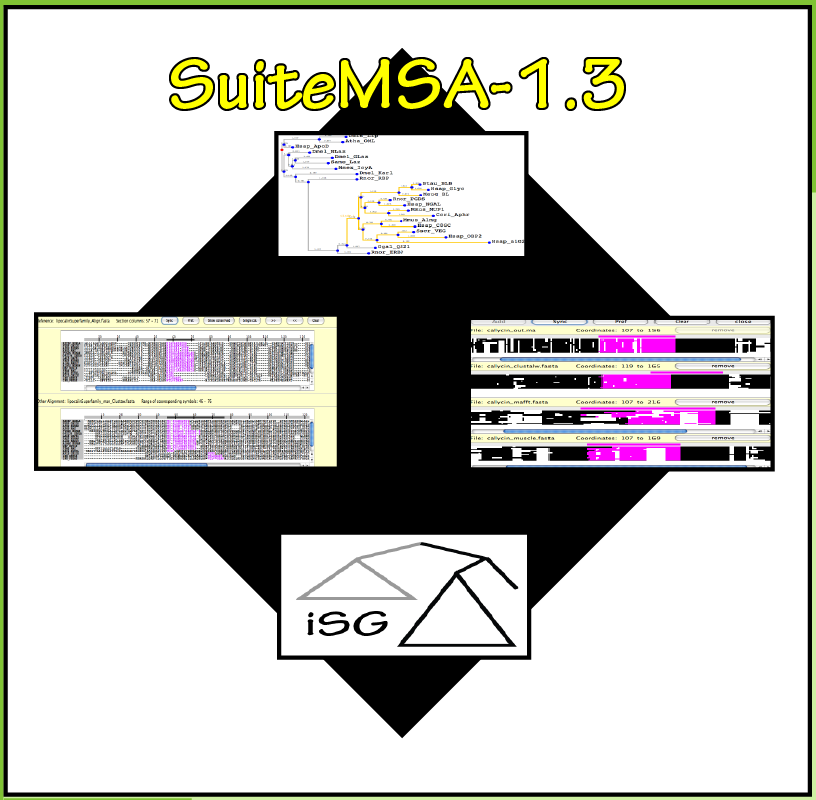
SuiteMSA
Visual Tools for Multiple Sequence Alignment Comparison and Molecular Sequence SimulationAnderson, C. L., Strope, C. L., and Moriyama, E. N. (2011) Assessing multiple sequence alignments using visual tools, in Bioinformatics: Trends and Methodologies (ed. Mahdavi, M. A.). InTech (ISBN 978-953-307-282-1)
Anderson, C. L., Strope, C. L. and Moriyama, E. N. (2011) SuiteMSA: Visual tools for multiple sequence alignment comparison and molecular sequence simulation. BMC Bioinformatics 12: 184

SimProDB
Simulated Protein Benchmark Database: large scale protein alignmentsCatherine Anderson (2017) Selecting the "closest to optimal" multiple sequence alignment using multi-layer perceptron. PhD dissertation, Department of Computer Science and Engineering, University of Nebraska—Lincoln.
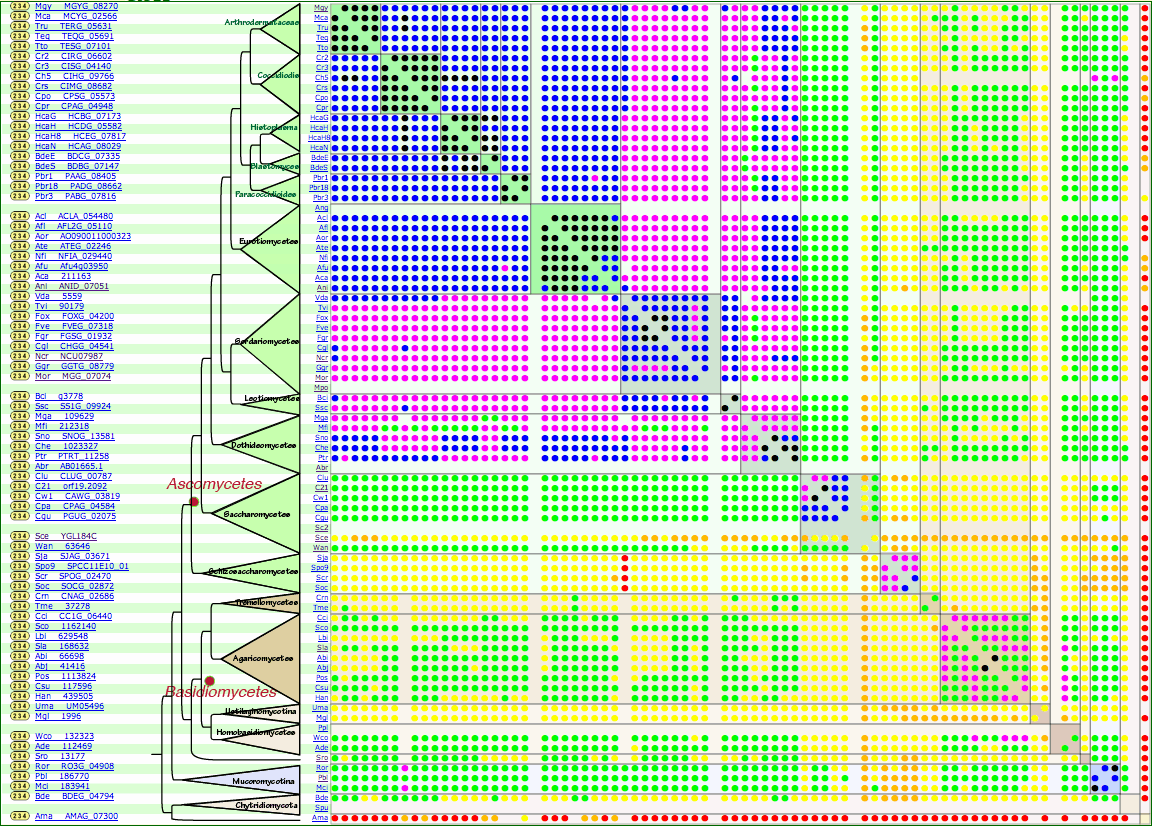
FGC
Fungal Genome Collection with Comparative Genomics InformationWilson, R. A., Fernandez, J., Quispe, C. F., Gradnigo, J., Seng, A., Moriyama, E. N., and Wright, J. D. (2012) Towards defining nutrient conditions encountered by the rice blast fungus during host infection. PLoS ONE 7: e47392
indel-Seq-Gen (iSG)
A protein family simulator that can incorporate domains, motifs, and indelsStrope, C. L., Scott, S. D. and Moriyama, E. N. (2007) indel-Seq-Gen: a new protein family simulator incorporating domains, motifs, and indels. Mol Biol Evol 24: 640-649.
Strope, C. L., Abel, K., Scott, S. D., and Moriyama, E. N. (2009) Biological sequence simulation for testing complex evolutionary hypotheses: indel-Seq-Gen version 2.0. Mol Biol Evol 26: 2581-2593.
iSGv2.1.0 Benchmark Datasets
Large and highly divergent sequence sets generated by iSGv2.1.0(last updated: June 10, 2018)