|
From Fig. 1, we can find MGG_07074 as the Magnaporthe oryzae orthologue.
Clicking on the MGG_07074 link in the orthologue matrix produces the MGG_07074 orthologue matrix (Fig. 2), which is very similar to Fig. 1
From these ortholog matrices, we can confirm that YGL184C in S. cerevisiae S288C and
MGG_07074 in M. oryzae are orthologues.
Orthologues in Aspergillus nidulans (ANID_07051, metG)
and in Neurospora crassa (NCU07987, met-2),
for example, can also be identified.
Note that many orthologues are highly similar to MGG_07074 (60% or higher).
In contrast, the S. cerevisiae orthologue (YGL184C) has much weaker similarity compared to others.
Yellow, orange, and red dots indicate that YGL184C has % identity lower than 60% against any other orthologues (Fig. 1 and Fig. 2)
|
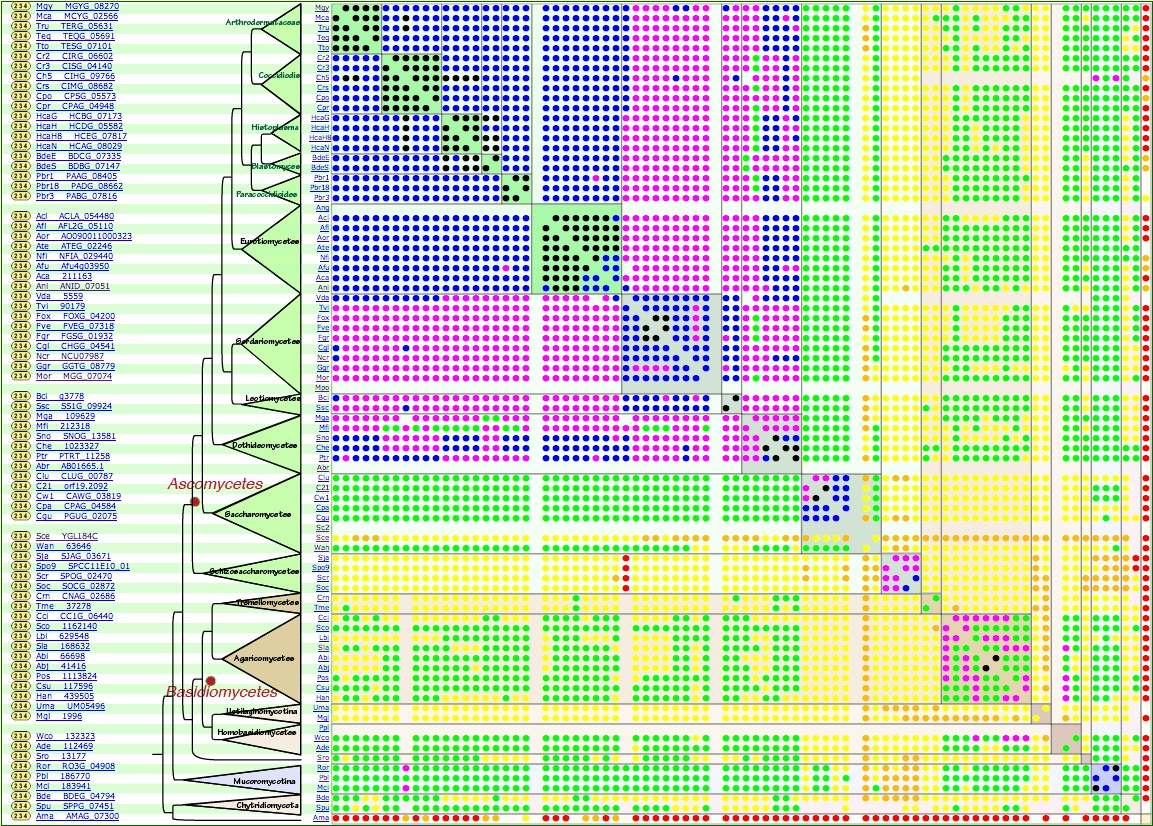
Fig. 2. (click the image to enlarge)
MGG_07074 orthologue distribution using 30% identity as the threshold.
|